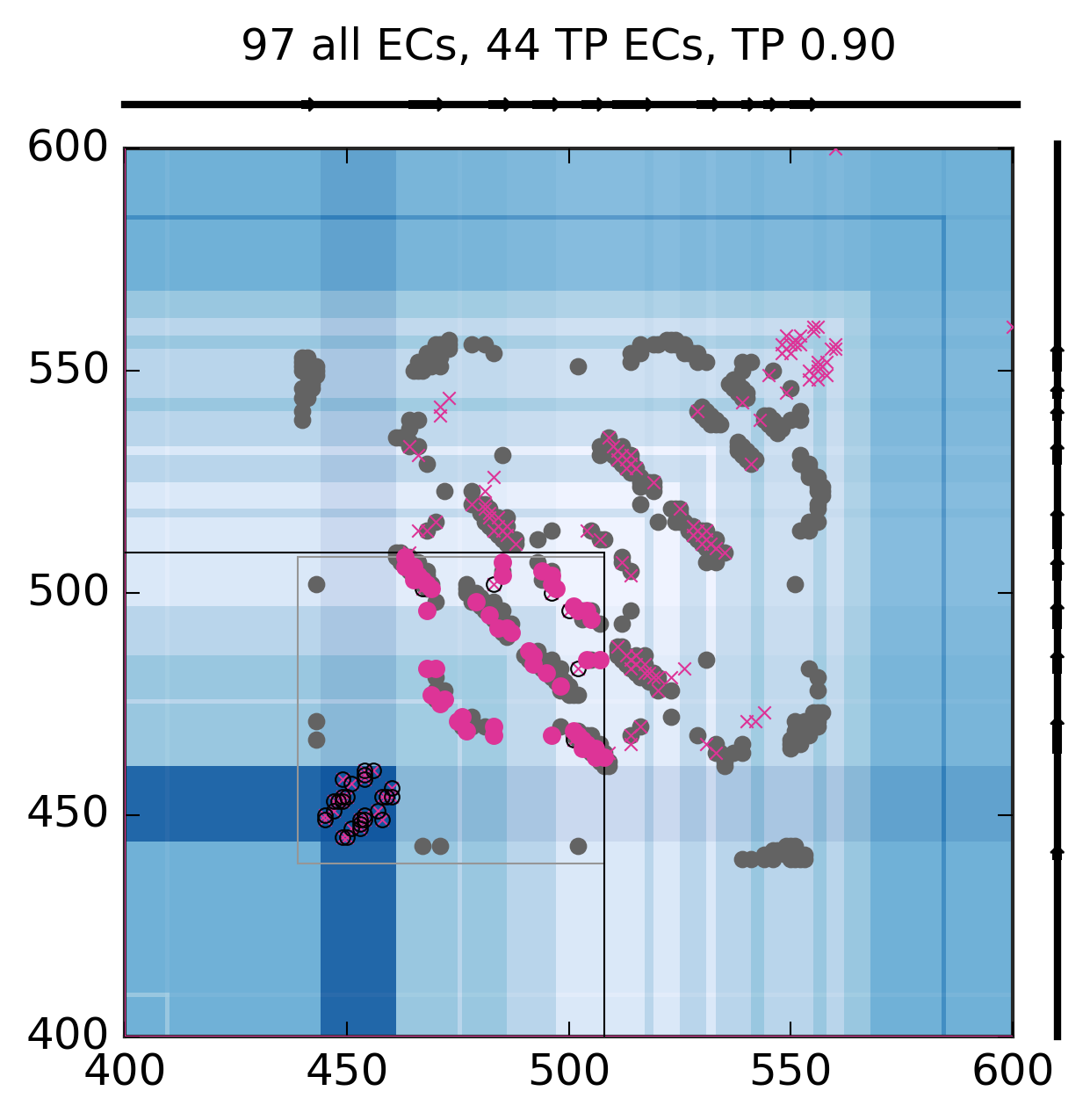
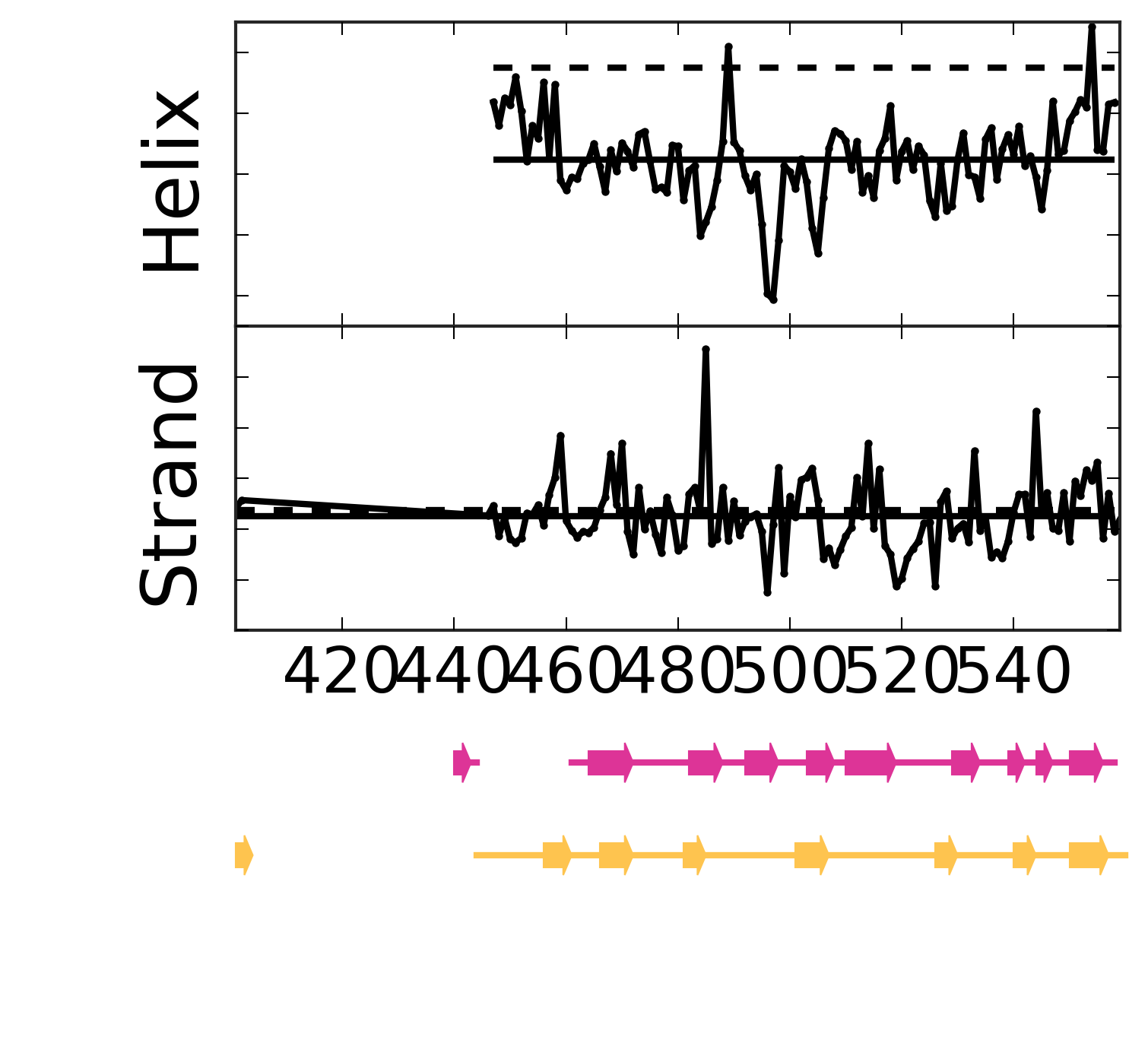
Evolutionary Coupling Analysis
Predicted and experimental contacts
Secondary structure from ECs
Known pdb structures
| pdb |
chain |
| 2x5p |
A |
| 4mli |
A |
| 4mls |
A |
EC score distribution and threshold
Top ECs
| Rank |
Residue 1 |
Amino acid 1 |
Residue 2 |
Amino acid 2 |
EC score |
| 1 |
482 |
T |
518 |
A |
2.30 |
| 2 |
486 |
R |
515 |
V |
1.88 |
| 3 |
484 |
E |
517 |
T |
1.82 |
| 4 |
515 |
V |
528 |
A |
1.59 |
| 5 |
486 |
R |
513 |
T |
1.57 |
| 6 |
467 |
K |
501 |
Q |
1.56 |
| 7 |
511 |
K |
532 |
T |
1.55 |
| 8 |
481 |
A |
520 |
P |
1.51 |
| 9 |
469 |
S |
477 |
E |
1.40 |
| 10 |
470 |
K |
516 |
E |
1.39 |
| 11 |
484 |
E |
515 |
V |
1.30 |
| 12 |
469 |
S |
501 |
Q |
1.27 |
| 13 |
468 |
F |
502 |
V |
1.25 |
| 14 |
513 |
T |
530 |
T |
1.19 |
| 15 |
511 |
K |
530 |
T |
1.19 |
| 16 |
467 |
K |
503 |
K |
1.16 |
| 17 |
513 |
T |
528 |
A |
1.09 |
| 18 |
485 |
L |
504 |
D |
1.08 |
| 19 |
552 |
I |
556 |
D |
0.94 |
| 20 |
465 |
H |
503 |
K |
0.92 |
| 21 |
487 |
D |
491 |
K |
0.89 |
| 22 |
519 |
A |
525 |
V |
0.85 |
| 23 |
479 |
A |
498 |
S |
0.84 |
| 24 |
496 |
W |
502 |
V |
0.84 |
| 25 |
510 |
G |
533 |
V |
0.84 |
| 26 |
484 |
E |
492 |
T |
0.83 |
| 27 |
485 |
L |
514 |
F |
0.81 |
| 28 |
514 |
F |
531 |
F |
0.81 |
| 29 |
456 |
M |
460 |
E |
0.81 |
| 30 |
486 |
R |
492 |
T |
0.78 |
| 31 |
482 |
T |
495 |
T |
0.77 |
| 32 |
481 |
A |
519 |
A |
0.76 |
| 33 |
471 |
R |
542 |
N |
0.73 |
| 34 |
465 |
H |
505 |
F |
0.73 |
| 35 |
466 |
I |
531 |
F |
0.70 |
| 36 |
468 |
F |
483 |
M |
0.68 |
| 37 |
463 |
A |
506 |
Y |
0.67 |
| 38 |
478 |
L |
520 |
P |
0.64 |
| 39 |
548 |
G |
556 |
D |
0.63 |
| 40 |
466 |
I |
514 |
F |
0.62 |
| 41 |
482 |
T |
517 |
T |
0.62 |
| 42 |
464 |
T |
509 |
P |
0.61 |
| 43 |
468 |
F |
514 |
F |
0.61 |
| 44 |
550 |
A |
556 |
D |
0.59 |
| 45 |
514 |
F |
529 |
I |
0.59 |
| 46 |
454 |
G |
458 |
I |
0.58 |
| 47 |
552 |
I |
558 |
Y |
0.58 |
| 48 |
472 |
D |
476 |
K |
0.57 |
| 49 |
468 |
F |
496 |
W |
0.57 |
| 50 |
497 |
I |
501 |
Q |
0.57 |
| 51 |
496 |
W |
504 |
D |
0.55 |
| 52 |
454 |
G |
459 |
E |
0.53 |
| 53 |
481 |
A |
523 |
Y |
0.53 |
| 54 |
471 |
R |
540 |
T |
0.52 |
| 55 |
509 |
P |
535 |
E |
0.52 |
| 56 |
470 |
K |
483 |
M |
0.51 |
| 57 |
550 |
A |
554 |
M |
0.51 |
| 58 |
465 |
H |
504 |
D |
0.51 |
| 59 |
483 |
M |
514 |
F |
0.50 |
| 60 |
485 |
L |
507 |
L |
0.50 |
| 61 |
465 |
H |
506 |
Y |
0.50 |
| 62 |
507 |
L |
512 |
Y |
0.49 |
| 63 |
488 |
S |
511 |
K |
0.49 |
| 64 |
556 |
D |
560 |
P |
0.48 |
| 65 |
464 |
T |
533 |
V |
0.48 |
| 66 |
529 |
I |
541 |
V |
0.47 |
| 67 |
504 |
D |
514 |
F |
0.46 |
| 68 |
548 |
G |
554 |
M |
0.46 |
| 69 |
549 |
D |
558 |
Y |
0.45 |
| 70 |
555 |
V |
560 |
P |
0.44 |
| 71 |
560 |
P |
600 |
Q |
0.43 |
| 72 |
445 |
G |
449 |
E |
0.43 |
| 73 |
512 |
Y |
531 |
F |
0.43 |
| 74 |
447 |
S |
453 |
S |
0.42 |
| 75 |
551 |
H |
557 |
A |
0.42 |
| 76 |
496 |
W |
500 |
G |
0.42 |
| 77 |
555 |
V |
559 |
K |
0.42 |
| 78 |
449 |
E |
454 |
G |
0.42 |
| 79 |
447 |
S |
451 |
G |
0.41 |
| 80 |
451 |
G |
457 |
T |
0.41 |
| 81 |
449 |
E |
458 |
I |
0.41 |
| 82 |
449 |
E |
453 |
S |
0.40 |
| 83 |
445 |
G |
450 |
Q |
0.40 |
| 84 |
471 |
R |
475 |
G |
0.40 |
| 85 |
483 |
M |
502 |
V |
0.40 |
| 86 |
473 |
I |
544 |
K |
0.40 |
| 87 |
539 |
V |
543 |
G |
0.40 |
| 88 |
545 |
A |
549 |
D |
0.39 |
| 89 |
483 |
M |
526 |
A |
0.39 |
| 90 |
463 |
A |
508 |
M |
0.39 |
| 91 |
551 |
H |
556 |
D |
0.39 |
| 92 |
448 |
S |
453 |
S |
0.39 |
| 93 |
549 |
D |
555 |
V |
0.39 |
| 94 |
494 |
S |
505 |
F |
0.38 |
| 95 |
466 |
I |
504 |
D |
0.38 |
| 96 |
454 |
G |
460 |
E |
0.38 |
| 97 |
450 |
Q |
454 |
G |
0.37 |
Alignment robustness analysis
First most common residue correlation
Second most common residue correlation